The Mediator complex: a central integrator of transcription
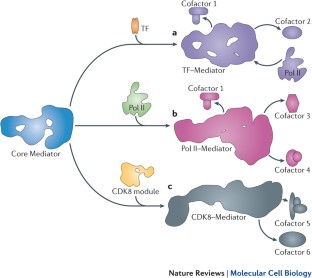
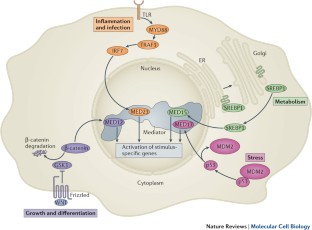
The RNA polymerase II (Pol II) enzyme transcribes all protein-coding and most non-coding RNA genes and is globally regulated by Mediator — a large, conformationally flexible protein complex with a variable subunit composition (for example, a four-subunit cyclin-dependent kinase 8 module can reversibly associate with it). These biochemical characteristics are fundamentally important for Mediator's ability to control various processes that are important for transcription, including the organization of chromatin architecture and the regulation of Pol II pre-initiation, initiation, re-initiation, pausing and elongation. Although Mediator exists in all eukaryotes, a variety of Mediator functions seem to be specific to metazoans, which is indicative of more diverse regulatory requirements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others

Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Article 02 August 2023
The Mediator complex as a master regulator of transcription by RNA polymerase II
Article 20 June 2022

Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases
Article 03 May 2022
References
- Plank, J. L. & Dean, A. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell55, 5–14 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Poss, Z. C., Ebmeier, C. C. & Taatjes, D. J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol.48, 575–608 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ansari, S. A. & Morse, R. H. Mechanisms of Mediator complex action in transcriptional activation. Cell. Mol. Life Sci.70, 2743–2756 (2013). ArticleCASPubMedGoogle Scholar
- Xu, W. & Ji, J. Y. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J. Genet. Genom.38, 439–452 (2011). ArticleCASGoogle Scholar
- Spaeth, J. M., Kim, N. H. & Boyer, T. G. Mediator and human disease. Semin. Cell Dev. Biol.22, 776–787 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Schiano, C., Casamassimi, A., Vietri, M. T., Rienzo, M. & Napoli, C. The roles of mediator complex in cardiovascular diseases. Biochim. Biophys. Acta1839, 444–451 (2014). ArticleCASPubMedGoogle Scholar
- Kulak, N. A., Pichler, G., Paron, I., Nagaraj, N. & Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nature Methods11, 319–324 (2014). ArticleCASPubMedGoogle Scholar
- Malik, S. et al. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol.24, 8244–8254 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Taatjes, D. J. & Tjian, R. Structure and function of CRSP/Med2: a promoter-selective transcriptional co-activator complex. Mol. Cell14, 675–683 (2004). ArticleCASPubMedGoogle Scholar
- Fondell, J. D., Ge, H. & Roeder, R. G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA93, 8329–8333 (1996). This paper reports the first biochemical purification of a human Mediator complex.ArticleCASPubMedPubMed CentralGoogle Scholar
- Stevens, J. L. et al. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science296, 755–758 (2002). ArticleCASPubMedGoogle Scholar
- Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B. & Roeder, R. G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell5, 683–693 (2000). ArticleCASPubMedGoogle Scholar
- D'Alessio, J. A., Ng, R., Willenbring, H. & Tjian, R. Core promoter recognition complex changes accompany liver development. Proc. Natl Acad. Sci. USA108, 3906–3911 (2011). ArticlePubMedPubMed CentralGoogle Scholar
- Deato, M. D. E. et al. MyoD targets TAF3/TRF3 to activate Myogenin transcription. Mol. Cell32, 96–105 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Herrera, F. J., Yamaguchi, T., Roelink, H. & Tjian, R. Core promoter factor TAF9B regulates neuronal gene expression. Elife3, e02559 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Liu, Y., Ranish, J. A., Aebersold, R. & Hahn, S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem.276, 7169–7175 (2001). ArticleCASPubMedGoogle Scholar
- Taatjes, D. J., Naar, A. M., Andel, F., Nogales, E. & Tjian, R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science295, 1058–1062 (2002). This study shows that the structural state of human Mediator changes upon binding to the activation domains of TFs.ArticleCASPubMedGoogle Scholar
- Knuesel, M. T., Meyer, K. D., Bernecky, C. & Taatjes, D. J. The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes Dev.23, 439–451 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Tsai, K. L. et al. A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nature Struct. Mol. Biol.20, 611–619 (2013). ArticleCASGoogle Scholar
- Davis, M. A. et al. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev.27, 151–156 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Barbieri, C. E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genet.44, 685–689 (2012). ArticleCASPubMedGoogle Scholar
- Li, N. et al. Cyclin C is a haploinsufficient tumour suppressor. Nature Cell Biol.16, 1080–1091 (2014). ArticleCASPubMedGoogle Scholar
- Makinen, N. et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science334, 252–255 (2011). ArticleCASPubMedGoogle Scholar
- Risheg, H. et al. A recurrent mutation in MED12 leading to R961W causes Opitz–Kaveggia syndrome. Nature Genet.39, 451–453 (2007). ArticleCASPubMedGoogle Scholar
- Ding, N. et al. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol. Cell31, 347–359 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Turunen, M. et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep.7, 654–660 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Donner, A. J., Ebmeier, C. C., Taatjes, D. J. & Espinosa, J. M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature Struct. Mol. Biol.17, 194–201 (2010). ArticleCASGoogle Scholar
- Galbraith, M. D. et al. HIF1A employs CDK8–Mediator to stimulate RNAPII elongation in response to hypoxia. Cell153, 1327–1339 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Alarcon, C. et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell139, 757–769 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bancerek, J. et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity38, 250–262 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hirst, M., Kobor, M. S., Kuriakose, N., Greenblatt, J. & Sadowski, I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell3, 673–678 (1999). ArticleCASPubMedGoogle Scholar
- Vincent, O. et al. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol.21, 5790–5796 (2001). ArticleCASPubMedPubMed CentralGoogle Scholar
- Morris, E. J. et al. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature455, 552–556 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhao, X. et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest.122, 2417–2427 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Daniels, D. L. et al. Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK–Mediator kinase module. J. Proteomics Bioinformhttp://dx.doi.org/10.4172/jpb.S2-004 (2013).
- Muncke, N. et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation108, 2843–2850 (2003). ArticleCASPubMedGoogle Scholar
- Mukhopadhyay, A. et al. CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Hum. Genet.128, 281–291 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Toth-Petroczy, A. et al. Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol.4, e1000243 (2008). This study reveals that yeast and human Mediator contain a preponderance of predicted IDRs.ArticleCASPubMedPubMed CentralGoogle Scholar
- Asturias, F. J., Jiang, Y. W., Myers, L. C., Gustafsson, C. M. & Kornberg, R. D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science283, 985–987 (1999). This paper provides the first insights into Mediator structure and the nature of the Mediator–Pol II interaction.ArticleCASPubMedGoogle Scholar
- Cai, G., Imasaki, T., Takagi, Y. & Asturias, F. A. Mediator structural conservation and implications for the regulation mechanism. Structure17, 559–567 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Meyer, K. D., Lin, S., Bernecky, C., Gao, Y. & Taatjes, D. J. p53 activates transcription by directing structural shifts in Mediator. Nature Struct. Mol. Biol.17, 753–760 (2010). This study gives structural and functional evidence that TF-induced structural shifts in Mediator are required for the activation of transcription.ArticleCASGoogle Scholar
- Naar, A. M., Taatjes, D. J., Zhai, W., Nogales, E. & Tjian, R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev.16, 1339–1344 (2002). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ebmeier, C. C. & Taatjes, D. J. Activator-Mediator binding regulates Mediator–cofactor interactions. Proc. Natl Acad. Sci. USA107, 11283–11288 (2010). ArticlePubMedPubMed CentralGoogle Scholar
- Tsai, K. L. et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell157, 1430–1444 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wang, X. et al. Redefining the modular organization of the core Mediator complex. Cell Res.24, 796–808 (2014). References 44 and 45 redefine previous models of yeast Mediator subunit architecture; reference 44 also provides the highest-resolution structure of yeast Mediator to date.ArticleCASPubMedPubMed CentralGoogle Scholar
- Dotson, M. R. et al. Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. USA97, 14307–14310 (2000). ArticleCASPubMedPubMed CentralGoogle Scholar
- Reeves, W. M. & Hahn, S. Activator-independent functions of the yeast Mediator Sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol.23, 349–358 (2003). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sakurai, H. & Fukasawa, T. Functional connections between mediator components and general transcription factors of Saccharomyces cerevisiae. J. Biol. Chem.275, 37251–37256 (2000). ArticleCASPubMedGoogle Scholar
- Sakurai, H., Kim, Y. J., Ohishi, T., Kornberg, R. D. & Fukasawa, T. The yeast GAL11 protein binds to the transcription factor IIE through GAL11 regions essential for its in vivo function. Proc. Natl Acad. Sci. USA93, 9488–9492 (1996). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ansari, S. A. et al. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J.31, 44–57 (2012). ArticleCASPubMedGoogle Scholar
- Lim, M. K. et al. Gal11p dosage-compensates transcriptional activator deletions via Taf14p. J. Mol. Biol.374, 9–23 (2007). ArticleCASPubMedGoogle Scholar
- Guglielmi, B. et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res.32, 5379–5391 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cevher, M. A. et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nature Struct. Mol. Biol.21, 1028–1034 (2014). This study demonstrates that an active, partial Mediator complex can be reconstituted from the recombinant expression of individual subunits and reveals key roles for MED14 and MED26.ArticleCASGoogle Scholar
- Imasaki, T. et al. Architecture of the Mediator head module. Nature475, 240–243 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lariviere, L. et al. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res.41, 9266–9273 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lariviere, L. et al. Structure of the Mediator head module. Nature492, 448–451 (2012). ArticleCASPubMedGoogle Scholar
- Robinson, P. J., Bushnell, D. A., Trnka, M. J., Burlingame, A. L. & Kornberg, R. D. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA109, 17931–17935 (2012). ArticlePubMedPubMed CentralGoogle Scholar
- Bernecky, C., Grob, P., Ebmeier, C. C., Nogales, E. & Taatjes, D. J. Molecular architecture of the human Mediator–RNA polymerase II–TFIIF assembly. PLoS Biol.9, e1000603 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Davis, J. A., Takagi, Y., Kornberg, R. D. & Asturias, F. A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell10, 409–415 (2002). ArticleCASPubMedGoogle Scholar
- Holstege, F. C. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell95, 717–728 (1998). This paper demonstrates a global requirement for Mediator and TFIIH for Pol II transcription in yeast.ArticleCASPubMedGoogle Scholar
- Myers, L. C., Gustafsson, C. M., Hayashibara, K. C., Brown, P. O. & Kornberg, R. D. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl Acad. Sci. USA96, 67–72 (1999). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kim, Y., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell77, 599–608 (1994). ArticleCASPubMedGoogle Scholar
- Myers, L. C. et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev.12, 45–54 (1998). ArticleCASPubMedPubMed CentralGoogle Scholar
- Thompson, C. M., Koleske, A. J., Chao, D. M. & Young, R. A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell73, 1361–1375 (1993). ArticleCASPubMedGoogle Scholar
- Guermah, M., Tao, Y. & Roeder, R. G. Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol. Cell. Biol.21, 6882–6894 (2001). ArticleCASPubMedPubMed CentralGoogle Scholar
- Johnson, K. M., Wang, J., Smallwood, A., Arayata, C. & Carey, M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev.16, 1852–1863 (2002). ArticleCASPubMedPubMed CentralGoogle Scholar
- Marr, M. T., Isogai, Y., Wright, K. J. & Tjian, R. Coactivator cross-talk specifies transcriptional output. Genes Dev.20, 1458–1469 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Takahashi, H. et al. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell146, 92–104 (2011). This study shows a direct role for MED26 in binding the SEC and affecting transcription elongation in human cells.ArticleCASPubMedPubMed CentralGoogle Scholar
- Xu, M. et al. Core promoter-selective function of HMGA1 and Mediator in Initiator-dependent transcription. Genes Dev.25, 2513–2524 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Baek, H. J., Kang, Y. K. & Roeder, R. G. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem.281, 15172–15181 (2006). ArticleCASPubMedGoogle Scholar
- Johnson, K. M. & Carey, M. Assembly of a mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr. Biol.13, 772–777 (2003). ArticleCASPubMedGoogle Scholar
- Jishage, M. et al. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol. Cell45, 51–63 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Esnault, C. et al. Mediator-dependent recruitment of TFIIH modules in Preinitiation Complex. Mol. Cell31, 337–346 (2008). ArticleCASPubMedGoogle Scholar
- Seizl, M., Lariviere, L., Pfaffeneder, T., Wenzeck, L. & Cramer, P. Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res.39, 6291–6304 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hu, X. et al. A Mediator-responsive form of metazoan RNA polymerase II. Proc. Natl Acad. Sci. USA103, 9506–9511 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cheng, B. et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell45, 38–50 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wu, Y. M. et al. Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-EM. EMBO J.31, 3575–3587 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Svejstrup, J. Q. et al. Evidence for a mediator cycle at the initiation of transcription. Proc. Natl Acad. Sci. USA94, 6075–6078 (1997). ArticleCASPubMedPubMed CentralGoogle Scholar
- Corden, J. L. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem. Rev.113, 8423–8455 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Eick, D. & Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev.113, 8456–8490 (2013). ArticleCASPubMedGoogle Scholar
- Boeing, S., Rigault, C., Heidemann, M., Eick, D. & Meisterernst, M. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. J. Biol. Chem.285, 188–196 (2010). ArticleCASPubMedGoogle Scholar
- Nair, D., Kim, Y. & Myers, L. C. Mediator and TFIIH govern carboxy-terminal domain-dependent transcription in yeast extracts. J. Biol. Chem.280, 33739–33748 (2005). ArticleCASPubMedGoogle Scholar
- Jeronimo, C. & Robert, F. Kin28 regulates the transient association of Mediator with core promoters. Nature Struct. Mol. Biol.21, 449–455 (2014). ArticleCASGoogle Scholar
- Wong, K. H., Jin, Y. & Struhl, K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell54, 601–612 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Rhee, H. S. & Pugh, B. F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature483, 295–301 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lee, S. K., Chen, X., Huang, L. & Stargell, L. A. The head module of Mediator directs activation of preloaded RNAPII in vivo. Nucleic Acids Res.41, 10124–10134 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science322, 1845–1848 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Seila, A. C. et al. Divergent transcription from active promoters. Science322, 1849–1851 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell130, 77–88 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Muse, G. W. et al. RNA polymerase is poised for activation across the genome. Nature Genet.39, 1507–1511 (2007). ArticleCASPubMedGoogle Scholar
- Zeitlinger, J. et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature Genet.39, 1512–1516 (2007). ArticleCASPubMedGoogle Scholar
- Kremer, S. B. et al. Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae. Genetics191, 95–106 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Malik, S., Wallberg, A. E., Kang, Y. K. & Roeder, R. G. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol.22, 5626–5637 (2002). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wang, G. et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell17, 683–694 (2005). ArticleCASPubMedGoogle Scholar
- Adelman, K. & Lis, J. T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature Rev. Genet.13, 720–731 (2012). ArticleCASPubMedGoogle Scholar
- Lin, C. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell37, 429–437 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Luo, Z., Lin, C. & Shilatifard, A. The super elongation complex (SEC) family in transcriptional control. Nature Rev. Mol. Cell Biol.13, 543–547 (2012). ArticleCASGoogle Scholar
- Paoletti, A. C. et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA103, 18928–18933 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Balamotis, M. A. et al. Complexity in transcription control at the activation domain–Mediator interface. Sci. Signal.2, ra20 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Park, J. M., Werner, J., Kim, J. M., Lis, J. T. & Kim, Y. J. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell8, 9–19 (2001). This is one of the first studies to suggest a role for Mediator in regulating paused or 'post-initiated' Pol II.ArticleCASPubMedGoogle Scholar
- Schaaf, C. A. et al. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet.9, e1003382 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kagey, M. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature467, 430–435 (2010). This study uncovers a role for Mediator in regulating genome architecture, implicating Mediator and cohesin in the formation and/or maintenance of enhancer–promoter gene loops in mammalian cells.ArticleCASPubMedPubMed CentralGoogle Scholar
- Lin, C., Garruss, A. S., Luo, Z., Guo, F. & Shilatifard, A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell152, 144–156 (2013). ArticleCASPubMedGoogle Scholar
- Guglielmi, B., Soutourina, J., Esnault, C. & Werner, M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl Acad. Sci. USA104, 16062–16067 (2007). ArticlePubMedPubMed CentralGoogle Scholar
- Kim, B. et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl Acad. Sci. USA104, 16068–16073 (2007). ArticlePubMedPubMed CentralGoogle Scholar
- Nock, A., Ascano, J. M., Barrero, M. J. & Malik, S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell48, 837–848 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Malik, S., Barrero, M. J. & Jones, T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl Acad. Sci. USA104, 6182–6187 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Larson, D. R. et al. Direct observation of frequency modulated transcription in single cells using light activation. ELife2, e00750 (2013). ArticlePubMedPubMed CentralGoogle Scholar
- Sandaltzopoulos, R. & Becker, P. B. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol.18, 361–367 (1998). ArticleCASPubMedPubMed CentralGoogle Scholar
- Yudkovsky, N., Ranish, J. A. & Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature408, 225–229 (2000). This study provides evidence for a re-initiation 'scaffold' form of PIC that contains most PIC factors, including Mediator.ArticleCASPubMedGoogle Scholar
- Reid, G. et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell11, 695–707 (2003). ArticleCASPubMedGoogle Scholar
- Andrau, J. et al. Genome-wide location of the coactivator Mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell22, 179–192 (2006). This is one of the first studies to reveal colocalization of Cdk8 with Mediator across the yeast genome.ArticleCASPubMedGoogle Scholar
- Mo, X., Kowenz-Leutz, E., Xu, H. & Leutz, A. Ras induces mediator complex exchange on C/EBPb. Mol. Cell13, 241–250 (2004). ArticleCASPubMedGoogle Scholar
- Kim, Y. K. et al. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J.25, 3596–3604 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pavri, R. et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell18, 83–96 (2005). This is the first study to reveal a potential means by which CDK8 module–Mediator interactions are regulated in mammalian cells.ArticleCASPubMedGoogle Scholar
- Ansari, S. A. et al. Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast. J. Biol. Chem.289, 14981–14995 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gilchrist, D. A. et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell143, 540–551 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Sharma, V. M., Li, B. & Reese, J. C. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev.17, 502–515 (2003). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lin, J. J. et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev.25, 2198–2209 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lemieux, K. & Gaudreau, L. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA polymerase II. EMBO J.23, 4040–4050 (2004). ArticleCASPubMedPubMed CentralGoogle Scholar
- Vilar, J. M. & Saiz, L. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev.15, 136–144 (2005). ArticleCASPubMedGoogle Scholar
- Levine, M., Cattoglio, C. & Tjian, R. Looping back to leap forward: transcription enters a new era. Cell157, 13–25 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Fanucchi, S., Shibayama, Y., Burd, S., Weinberg, M. S. & Mhlanga, M. M. Chromosomal contact permits transcription between coregulated genes. Cell155, 606–620 (2013). ArticleCASPubMedGoogle Scholar
- Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell148, 84–98 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell149, 1233–1244 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kieffer-Kwon, K. R. et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell155, 1507–1520 (2013). ArticleCASPubMedGoogle Scholar
- Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature489, 109–113 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell153, 1281–1295 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell155, 934–947 (2013). ArticleCASPubMedGoogle Scholar
- Muto, A. et al. Nipbl and Mediator cooperatively regulate gene expression to control limb development. PLoS Genet.10, e1004671 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Dobi, K. C. & Winston, F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol.27, 5575–5586 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Mukundan, B. & Ansari, A. Srb5/Med18-mediated termination of transcription is dependent on gene looping. J. Biol. Chem.288, 11384–11394 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Orom, U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell143, 46–58 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature494, 497–501 (2013). This study demonstrates that ncRNAs transcribed from distal sequence elements can interact with Mediator to regulate gene looping to the promoter and transcription activation.ArticleCASPubMedPubMed CentralGoogle Scholar
- Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature474, 390–394 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hah, N., Murakami, S., Nagari, A., Danko, C. G. & Kraus, W. L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res.23, 1210–1223 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lam, M. T. et al. Rev–Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature498, 511–515 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature498, 516–520 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hsieh, C. L. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl Acad. Sci. USA111, 7319–7324 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Step, S. E. et al. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARγ-driven enhancers. Genes Dev.28, 1018–1028 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bartolomei, M. S., Halden, N. F., Cullen, C. R. & Corden, J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol.8, 330–339 (1988). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gerber, H. P. et al. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature374, 660–662 (1995). ArticleCASPubMedGoogle Scholar
- Oya, E. et al. Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet.9, e1003677 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Carlsten, J. O. et al. Mediator promotes CENP-a incorporation at fission yeast centromeres. Mol. Cell. Biol.32, 4035–4043 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Thorsen, M., Hansen, H., Venturi, M., Holmberg, S. & Thon, G. Mediator regulates non-coding RNA transcription at fission yeast centromeres. Epigenetics Chromatin5, 19 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lenstra, T. L. et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell42, 536–549 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Peng, J. & Zhou, J. Q. The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance. Nucleic Acids Res.40, 581–593 (2012). ArticleCASPubMedGoogle Scholar
- Suzuki, Y. & Nishizawa, M. The yeast GAL11 protein is involved in regulation of the structure and the position effect of telomeres. Mol. Cell. Biol.14, 3791–3799 (1994). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhu, X. et al. Mediator influences telomeric silencing and cellular life span. Mol. Cell. Biol.31, 2413–2421 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Marr, S. K., Lis, J. T., Treisman, J. E. & Marr, M. T. The metazoan-specific Mediator subunit 26 (Med26) is essential for viability and is found at both active genes and pericentric heterochromatin in Drosophila. Mol. Cell. Biol.34, 2710–2720 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Smallwood, A., Black, J. C., Tanese, N., Pradhan, S. & Carey, M. HP1-mediated silencing targets Pol II coactivator complexes. Nature Struct. Mol. Biol.15, 318–320 (2008). ArticleCASGoogle Scholar
- Jiang, Y. W. et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl Acad. Sci. USA95, 8538–8543 (1998). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gaarenstroom, T. & Hill, C. S. TGF-β signaling to chromatin: how SMADs regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol.32, 107–118 (2014). ArticleCASPubMedGoogle Scholar
- Kato, Y., Habas, R., Katsuyama, Y., Naar, A. & He, X. A component of the ARC/Mediator complex required for TGF β/Nodal signalling. Nature418, 641–646 (2002). ArticleCASPubMedGoogle Scholar
- Zhao, M. et al. Mediator MED15 modulates transforming growth factor β (TGFβ)/Smad signaling and breast cancer cell metastasis. J. Mol. Cell. Biol.5, 57–60 (2013). ArticlePubMedGoogle Scholar
- Huang, S. et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell151, 937–950 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Knuesel, M. T., Meyer, K. D., Donner, A. J., Espinosa, J. M. & Taatjes, D. J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of Mediator. Mol. Cell. Biol.29, 650–661 (2009). ArticleCASPubMedGoogle Scholar
- Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell36, 457–468 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kahn, M. Can we safely target the WNT pathway? Nature Rev. Drug Discov.13, 513–532 (2014). ArticleCASGoogle Scholar
- Carrera, I., Janody, F., Leeds, N., Duveau, F. & Treisman, J. E. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl Acad. Sci. USA105, 6644–6649 (2008). ArticlePubMedPubMed CentralGoogle Scholar
- Rocha, P. P., Scholze, M., Bleiss, W. & Schrewe, H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development137, 2723–2731 (2010). ArticleCASPubMedGoogle Scholar
- Kim, S., Xu, X., Hecht, A. & Boyer, T. G. Mediator is a transducer of Wnt/β-catenin signaling. J. Biol. Chem.281, 14066–14075 (2006). ArticleCASPubMedGoogle Scholar
- Zhang, H. & Emmons, S. W. A C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes Dev.14, 2161–2172 (2000). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhao, J., Ramos, R. & Demma, M. CDK8 regulates E2F1 transcriptional activity through S375 phosphorylation. Oncogene32, 3520–3530 (2012). ArticleCASPubMedGoogle Scholar
- Firestein, R. et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature455, 547–551 (2008). References 33 and 165 identifyCDK8as a colon cancer oncogene.ArticleCASPubMedPubMed CentralGoogle Scholar
- Kasza, A. Signal-dependent Elk-1 target genes involved in transcript processing and cell migration. Biochim. Biophys. Acta1829, 1026–1033 (2013). ArticleCASPubMedGoogle Scholar
- Wang, W. et al. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev. Cell16, 764–771 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Yin, J. W. et al. Mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes Dev.26, 2192–2205 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Belakavadi, M., Pandey, P. K., Vijayvargia, R. & Fondell, J. D. MED1 phosphorylation promotes its association with Mediator: implications for nuclear receptor signaling. Mol. Cell. Biol.28, 3932–3942 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pandey, P. K. et al. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell. Biol.25, 10695–10710 (2005). ArticleCASPubMedPubMed CentralGoogle Scholar
- Papantonis, A. & Cook, P. R. Transcription factories: genome organization and gene regulation. Chem. Rev.113, 8683–8705 (2013). ArticleCASPubMedGoogle Scholar
- Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell149, 768–779 (2012). ArticleCASPubMedGoogle Scholar
- Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell149, 753–767 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Tompa, P. Hydrogel formation by multivalent IDPs: a reincarnation of the microtrabecular lattice? Intrinsically Disordered Proteins1, e24068 (2013). ArticlePubMedPubMed CentralGoogle Scholar
- Takagi, Y. & Kornberg, R. D. Mediator as a general transcription factor. J. Biol. Chem.281, 80–89 (2006). ArticleCASPubMedGoogle Scholar
- Bourbon, H. M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res.36, 3993–4008 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev.114, 6589–6631 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Madhani, H. D. The frustrated gene: origins of eukaryotic gene expression. Cell155, 744–749 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kuuluvainen, E. et al. Cyclin-dependent kinase 8 module expression profiling reveals requirement of Mediator subunits 12 and 13 for transcription of Serpent-dependent innate immunity genes in Drosophila. J. Biol. Chem.289, 16252–16261 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Napoli, C., Sessa, M., Infante, T. & Casamassimi, A. Unraveling framework of the ancestral Mediator complex in human diseases. Biochimie94, 579–587 (2012). ArticleCASPubMedGoogle Scholar
- Phillips, A. J. & Taatjes, D. J. Small molecule probes to target the human Mediator complex. Isr. J. Chem.53, 588–595 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Milbradt, A. G. et al. Structure of the VP16 transactivator target in the Mediator. Nature Struct. Mol. Biol.18, 410–415 (2011). ArticleCASGoogle Scholar
- Vojnic, E. et al. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nature Struct. Mol. Biol.18, 404–409 (2011). ArticleCASGoogle Scholar
- Yang, F. et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature442, 700–704 (2006). ArticleCASPubMedGoogle Scholar
- Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell153, 320–334 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell153, 307–319 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science330, 1775–1787 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science330, 1787–1797 (2010).
- Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. Locus control regions. Blood100, 3077–3086 (2002). ArticleCASPubMedGoogle Scholar
- Darnell, J. E. Jr. Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA19, 443–460 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Grunberg, S. & Hahn, S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem. Sci.38, 603–611 (2013). ArticleCASPubMedGoogle Scholar
- Thomas, M. C. & Chiang, C. M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol.41, 105–178 (2006). ArticleCASPubMedGoogle Scholar
- He, Y., Fang, J., Taatjes, D. J. & Nogales, E. Structural visualization of key steps in human transcription initiation. Nature495, 481–486 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Murakami, K. et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science342, 1238724 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Muhlbacher, W. et al. Conserved architecture of the core RNA polymerase II initiation complex. Nature Commun.5, 4310 (2014). ArticleCASGoogle Scholar
- Bieniossek, C. et al. The architecture of human general transcription factor TFIID core complex. Nature493, 699–702 (2013). ArticleCASPubMedGoogle Scholar
- Bernecky, C. & Taatjes, D. J. Activator–Mediator binding stabilizes RNA polymerase II orientation within the human Mediator–RNA polymerase II–TFIIF assembly. J. Mol. Biol.417, 387–394 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Elmlund, H. et al. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. USA103, 15788–15793 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
Acknowledgements
The authors thank A. Shilatifard and the reviewers for comments on the manuscript. They apologize that they could not discuss other important and relevant research owing to space and citation limits. D.J.T.'s laboratory is supported by the US National Science Foundation (MCB-1244175) and the US National Cancer Institute (CA175849; CA1707041; CA175448). B.L.A. has been supported in part by the US National Institutes of Health (T32 GM08759).








